|
parameters.h File Reference IntroductionDefinition of the functions to deal with the parameters given in a .param file. The available parameters are:
Note that internal names for parameters are preceded by CT_, so, for instance EPSILON is called CT_EPSILON in the code
Definition in file parameters.h.
Macro Definition Documentation◆ REP_LINKS
In this representation we use 6 parameters to represent two vectors of the rotation matrices for each link. The third vector is the cross product of the two represented vectors.
Definition at line 111 of file parameters.h. ◆ REP_FLINKS
In this representation we use 9 parameters to represent the full rotation matrices for each link.
Definition at line 121 of file parameters.h. ◆ REP_QLINKS
In this representation we use 4 parameters to represent rotation matrices for each link relying on quaternions.
Definition at line 132 of file parameters.h. ◆ REP_JOINTS
In this representation we do not use any variable for links but only for joints. This is similar to the classical DH non redundant formulation, but includes some auxliary/dummy variables for some kind of joints (in_patch).
Definition at line 144 of file parameters.h. ◆ SOLID
◆ VCOLLIDE
◆ PQP
◆ FCL
◆ C_FCL
◆ BULLET
◆ C_BULLET
◆ RIGIDCLL
◆ NOCD
◆ AMBIENT_SAMPLING
In this sampling mode, points are generated randomly in the ambient space box.
Definition at line 235 of file parameters.h. ◆ KDTREE_SAMPLING
In this sampling mode, points are generated randomly in the boxes bounding the tree nodes. These boxes are defined in the kd-tree.
Definition at line 246 of file parameters.h. ◆ TANGENT_SAMPLING
In this sampling mode, points are generated randomly in the tangent spaces stored in the charts. This is only used in AtlasRRT. If the sampling mode is set to this value normal RRTs use AMBIENT_SAMPLING
Definition at line 258 of file parameters.h. ◆ DEFAULT_PARAMS
Default file of parameters. This is the file of parameters read in the first place, before reading the file specific for the problem at hand. In this way, the problem specific parameter need only to include values for the parameters whose value is not to be the default value. Definition at line 269 of file parameters.h. ◆ CT_EPSILON
Threshold to turn any number into 0. For position analysis problmes (relying on a linear program) it should be larger than 1e-6, the threshold hard-coded in many simplex packages. For path/motion planning we typically use values of 1e-8 or even 1e-10, depending on the accuracy of the given initial and final configurations/states. Definition at line 281 of file parameters.h. ◆ CT_REPRESENTATION
The mechanisms configurations can be represented in different ways:
When using JOINTS representation configurations/states are read form a .joints file. Otherwise they are read form a .links file. Note that if you change this parameter solutions files previosly computed can become invalid. This can be a source of confusion so care must be taken to create/manipulate solutions using the same value for this parameter. Definition at line 305 of file parameters.h. ◆ CT_RHO
If boxes in a prune process reduce more that RHO, the reduction process is repeated. Definition at line 312 of file parameters.h. ◆ CT_SMALL_SIGMA
A prune process that reduces the size of a box is stoppes when the box reaches this size. This is typically the size of the boxes bounding isolated solutions. Definition at line 320 of file parameters.h. ◆ CT_SIGMA
If at the beginning of a prune process the box is smaller than this size, it is already considered a solution. Typically this is the size of the boxes bounding continuous solution sets. Definition at line 329 of file parameters.h. ◆ CT_E
Maximum error between the manifold and the tangent space for each chart. Definition at line 336 of file parameters.h. ◆ CT_CE
Maximum oriented curvature error of the local maps for each chart. Definition at line 343 of file parameters.h. ◆ CT_R
Maximum radius of the local maps when building atlas (or part of atlas). Definition at line 350 of file parameters.h. ◆ CT_SR
Radius of the ball used to generate samples. This must be larger than CT_R.
Definition at line 359 of file parameters.h. ◆ CT_DELTA
Step size for the paths connecting two given configurations. In between two steps no collision detetion is performed so, indirectly, this parameter defines the maximum penetrability between any two bodies in the problem. Definition at line 368 of file parameters.h. ◆ CT_ATLASGBF_BETA
Cost penalty factor for cuikatlasGBF. The cuikatlasGBF was our first planning algorihtm using higher-dimensional continuation but we hardly use it nowadays since the cuikatlasrrt typically performs better (spacially in high dimensions). See this paper for more details. Definition at line 379 of file parameters.h. ◆ CT_STATE_PERIOD
Period between two consecutive savings of the internal cuik state. If the computation is interrupted, the solving process is re-started from the last saved state. Definition at line 388 of file parameters.h. ◆ CT_N_SOLUTIONS
Number of output solutions boxes we need. Use 0 to obtain all possible solution boxes for the problem at hand. Definition at line 396 of file parameters.h. ◆ CT_MAX_NEWTON_ITERATIONS
Maximum number iterations in the Newton-Raphson function. This is used, for instance, to implement the tangent-to-manifold map used in the chart implementation. Definition at line 405 of file parameters.h. ◆ CT_N_DOF
Dimensionality of the solution space for the mechanism at hand. If set to 0, the system tries to guess this value automatically assuming that the first point in the joints/links file is regular. Definition at line 415 of file parameters.h. ◆ CT_GAMMA
Contant part of the search radius for nearest neightours in RRT*. See this paper for details. Definition at line 423 of file parameters.h. ◆ CT_DUMMIFY
Dummification level.
Definition at line 437 of file parameters.h. ◆ CT_SPLIT_TYPE
Select the criterion used to determine the split dimension for a box in a branch-and-prune process:
Definition at line 447 of file parameters.h. ◆ CT_SAFE_SIMPLEX
Trade off between speed and numerical stability when using the simplex Possible values are
Definition at line 459 of file parameters.h. ◆ CT_SIMPLIFICATION_LEVEL
Simplification level:
The less the simplification the more the numerical stability, but the slower the solution process. A simplification level 2 should be Ok except for extremely ill conditioned cases. Definition at line 479 of file parameters.h. ◆ CT_LR2TM_Q
Threshold to switch from a specific linearization (one based on linear relaxations) to a general one (based on Taylor models) for purely quadratic equations: parabolas, circles, and spheres. Definition at line 490 of file parameters.h. ◆ CT_LR2TM_S
Threshold to switch from a specific linearization (one based on linear relaxations) to a general one (based on Taylor models) for saddle equations. Definition at line 499 of file parameters.h. ◆ CT_VDW_RATIO
Reduction ratio [0,1] over the Van der Waals radius used to detect steric clashes. The reduction is used to ensure that only very high energy conformations are discarded by considering the collisions between atoms (as if atoms where solid spheres). Definition at line 509 of file parameters.h. ◆ CT_CUT_X
Limit of domain of the X dimension of 3d plots when the variables have circular ranges. The default limits are [-pi,pi] and this sometimes produce cutted plots. Using these constants the range of the plot can be adjusted at will to avoid those cuts. This is basically used when plotting atlas the DOF CT_REPRESENTATION. Definition at line 521 of file parameters.h. ◆ CT_CUT_Y
Limit of domain of the Y dimension of 3d plots when the variables have circular ranges. The default limits are [-pi,pi] and this sometimes produce cutted plots. Using these constants the range of the plot can be adjusted at will to avoid those cuts. This is basically used when plotting atlas the DOF CT_REPRESENTATION. Definition at line 533 of file parameters.h. ◆ CT_CUT_Z
Limit of domain of the Z dimension of 3d plots when the variables have circular ranges. The default limits are [-pi,pi] and this sometimes produce cutted plots. Using these constants the range of the plot can be adjusted at will to avoid those cuts. This is basically used when plotting atlas the DOF CT_REPRESENTATION. Definition at line 545 of file parameters.h. ◆ CT_COEF_TEMP
Scale factor of the temperature parameter for Transition-based RRT. This is part of our implementation of the Transition-based RRT that we used in this paper. For more details on the Transition-based RRT see this paper. Definition at line 556 of file parameters.h. ◆ CT_NFAIL_MAX
Number of successive trnasition test failues requiered before increasing the temperature. This is part of our implementation of the Transition-based RRT that we used in this <a href="http://www.iri.upc.edu/people/porta/Docs/jcc13.pdf</a>. For more details on the Transition-based RRT see this <a href="https://ieeexplore.ieee.org/document/4650993">paper. Definition at line 569 of file parameters.h. ◆ CT_DETECT_BIFURCATIONS
To activate/deactivate the search for bifurcations during the atlas construction. See this report for more details about branch switching. Definition at line 580 of file parameters.h. ◆ CT_MAX_PLANNING_TIME
Maximum time in seconds for the path planning algorihtms (cuikatlasGBF, cuikaltasrrt, cuikccrrt,....). Definition at line 587 of file parameters.h. ◆ CT_MAX_PLANNING_ITERATIONS
Maximum iterations for the path planning algorihtms (currently only used in AtlasRRT* and RRT* since the rest of algorithms are limited in time only). Definition at line 595 of file parameters.h. ◆ CT_MAX_CHARTS
Maximum number of charts in at atlas. Definition at line 602 of file parameters.h. ◆ CT_MAX_NODES_RRT
Maximum number of nodes in an RRT. Definition at line 609 of file parameters.h. ◆ CT_BI_RRT
TRUE if the RRT search is bi-directional. cuikccrrt is one-directional despite the value of this parameter. Definition at line 620 of file parameters.h. ◆ CT_RRT_GRAPH
TRUE if the RRT has to store all the neighbours for each new node. This is only used in cuikatlasrrtstar to speed up the propagation of improvements of the path to the nodes. See this paper for more details. Definition at line 632 of file parameters.h. ◆ CT_DYNAMIC_DOMAIN
TRUE if the RRT-like planners have to use dynamic domains. See this paper for more details on this technique. Definition at line 642 of file parameters.h. ◆ CT_CD_ENGINE
◆ CT_SAMPLING
Parameter used to select the mode to generate samples to expand the (Atlas)RRT. Three modes are available:
See this report for more details. Definition at line 675 of file parameters.h. ◆ CT_PRETENSION
Norm of the tension/compression forces in the tensegrity. In equilibrium, these forces can be arbitrarily scaled preserving the equilibrium. By imposing a norm to the forces this scale factor (a short of useless degree of freedom) is cancelled. If set to 0, the scale factor is not fixed. Note that in general, the equilibrium forces can have more than one degree of freedom (the kernel of the equilibrium matrix can be larger than one). We only cancel one degree of fredom. If there are more, they can be cancelled by fixing some tensions or compressions to a constant value. See this paper for more details. Definition at line 693 of file parameters.h. ◆ CT_DYNAMICS
If larger than 0, if the dynamics must be considered. If so, the problems are defined in phase space (position and velocity) where the velocity variables and equations are automatically generated. Moreover, when de dynamics is considered, the (Atlas)RRT is extended integrating the dynamic equation. The value (>0) is used to select the integration method:
Definition at line 715 of file parameters.h. ◆ CT_INTEGRATION_TIME
When considering dynamics, this fixes the integration time for each selected action. Definition at line 723 of file parameters.h. ◆ CT_DEFAULT_MAX_VELOCITY
The maximum velocity (in absolute value). Only used if the maximum velocity can not be taken from the world file. Definition at line 732 of file parameters.h. ◆ CT_N_DYNAMIC_ACTIONS
Number of valid actions when performing dynamic simulations/planning. Typically, such actions are samples in a continuous domain of valid forces/torques which lead to valid accelerations. If set to 0, an LQR-based method is used to determine the action to execute. Definition at line 745 of file parameters.h. ◆ CT_DYNAMIC_GOAL_ERROR
Tolerance when aiming at a goal state (or the connection between thress) in a problem with dynamics. This is call beta in our papers on kinodynamic planning. Not to be confused with ATLASGBF_BETA. Definition at line 757 of file parameters.h. ◆ CT_G_COMPENSATION
If set, actions are interpreted as variations over the action that compensates the gravity effects. Definition at line 765 of file parameters.h. ◆ CT_NEG_LM
If set the Lagrange multipliers should be negative for a state to be valid. So far this is only used in cable-driven robots, to ensure that cables are in tension since the sign of the Lagrange multipliers is related with the sign of the internal forces in the mechanisms. See this <a href="http://www.iri.upc.edu/people/porta/Docs/cc17.pdf"paper for more details. Definition at line 777 of file parameters.h. ◆ CT_G_AXIS
Select the gravity axis: 0 for no gravity, 1 for X, 2 for Y, 3 for Z, 4 for -X, 5 for -Y and 6 for Z Definition at line 785 of file parameters.h. ◆ CT_TAU
Balance between the relevance of time and the relevance of the action effort when computing the optimal action via LQR. Definition at line 793 of file parameters.h. ◆ CT_LQR_PLANNING_TIME
This parameter determines the planning time when using LQR to determine the policy. It is typically related to CT_INTEGRATION_TIME Definition at line 801 of file parameters.h. ◆ NPARAMETERS
Total number of parameters. Definition at line 808 of file parameters.h. Typedef Documentation◆ Tparameters
A set of parameters. Each parameter is identified with a constant defined in file parameters.h. This is basically a set of constants, but only a small, predefined set of constants are allowed. This is why we do not re-use Tconstants but provide a different implementation.
Definition at line 843 of file parameters.h. Function Documentation◆ InitParameters()
Defines an empty set of parameters.
Definition at line 36 of file parameters.c. References NPARAMETERS. Referenced by InitParametersFromFile(). ◆ InitParametersFromFile()
Defines an set of parameters from a file. It first read the default set of parameters and then the given set of parameters.
Definition at line 51 of file parameters.c. References CreateFileName(), DEFAULT_PARAMS, DeleteFileName(), Error(), GetFileFullName(), InitParameters(), NPARAMETERS, PARAM_EXT, ReadParameters(), and Warning(). Referenced by main(). ◆ GetParameter()
Gets the value for a particular parameter.
Definition at line 101 of file parameters.c. References Error(), and NPARAMETERS. Referenced by AddBranchToAtlasDynamicRRT(), AddBranchToAtlasRRT(), AddBranchToRRT(), AddNodeToRRT(), AddStepToAtlasRRTstar(), AddStepToRRTstar(), AddTrustedChart2Atlas(), AddVelocityEquations(), ApplyLinkRot(), ApplyLinkRotVar(), ApproachState(), AtlasAStar(), AtlasBiRRTstar(), AtlasGBF(), AtlasRRT(), AtlasRRTSimulate(), AtlasRRTstar(), AtlasTRRT(), BiRRTstar(), BuildAtlasFromPoint(), cBiRRT(), ccRRT(), ccTRRT(), Chart2Manifold(), ClassifyPointInChart(), CombineGradients(), ComputeAcceleration(), ComputeHandC_FJH(), ComputeInverseDynamics(), ComputeSplitDimInt(), ConnectDynamicStates(), ConnectDynamicStatesID(), ConnectSamples(), ConnectSamplesChart(), CSRemoveLCVars(), CSRemoveUnusedVars(), CSRemoveVarsWithCtRange(), CuikGradientInBox(), CuikNewtonInBox(), CuikNewtonSimp(), DealWithCP(), DetermineChartNeighbours(), DistanceOnChart(), DummifyAndAddEquation(), ExtendAtlasFromPoint(), ExtendAtlasTowardPoint(), FindSingularPoint(), FixLinks(), FixLinkZToZero(), FixZToZero(), GenerateEquationsFromBranch(), GenerateForceEquilibriumEquations(), GenerateJointEquations(), GenerateJointEquationsInBranch(), GenerateJointRangeEquations(), GenerateJointRangeSingularityEquations(), GenerateJointSolution(), GenerateLinkConf(), GenerateLinkRot(), GenerateLinkSolution(), GenerateTransEquationsFromBranch(), GenerateWorldEquations(), GenerateWorldSingularityEquations(), GeodesicDistance(), GetChartDegree(), GetJointTransSeq(), GetLinkPoseSimpVars(), GetLinkTransformsFromSolution(), GetLinkTransformsFromSolutionPoint(), GetSCpSystem(), GetSolutionPointFromLinkTransforms(), GetTransform2Link(), GradientSmooth(), IncrementalSampleCuikSystemInBox(), InitAtlasFromPoint(), InitAtlasRRT(), InitBranchState(), InitChartInt(), InitDynamicSpace(), InitHandC(), InitRRT(), InitWorldCD(), InitWorldFromMolecule(), InitWorldKinCS(), kinobiRRT(), kinoEST(), kinoRRT(), LinearizeDynamics(), LQRComputePolicy(), LQRComputePolicy_t(), main(), ManifoldDimension(), MinimizeOnAtlas(), MoveWorld(), MPI_SolveCuikSystem(), NewChartFromPoint(), NewTemptativeSample(), Newton2ManifoldPlane(), NextDynamicState(), NextDynamicStateEuler(), NextDynamicStateLocalEuler(), NextDynamicStateLocalRK4(), NextDynamicStateRK4(), NumericIntegration(), PathEffort(), PathInChart(), PathStart2GoalInRRT(), PlotAtlasRRT(), PlotChart(), PlotChartAsPolygon(), PlotForceField(), PlotRRT(), PlotSamples(), PointOnChart(), Polytope2SPolytope(), PopulateWithSamples(), PostProcessBox(), PrintAtlasStatistics(), PrintCuikSystemWithSimplification(), PrintWorldAxes(), RandomSmooth(), ReadOneSample(), ReadTwoSamples(), RecursiveReWireRRTstar(), ReduceBox(), ReduceBoxEquationWise(), RefineSingularPoint(), RefineTrajectory(), RegenerateJointBox(), RegenerateJointSolution(), RegenerateLinkBox(), RegenerateLinkSolution(), RegenerateMechanismBox(), RegenerateSolution(), RegenerateWorldOriginalPoint(), ReWireAtlasRRTstar(), ReWireRRTstar(), RRTstar(), SampleCuikSystemInBox(), ShortcutSmooth(), SimplifyCuikSystem(), Simulate(), SmoothSamples(), SolveCuikSystem(), StepCostNumericalGradient(), StepDispersionGradient(), StepEffort(), StepEffortGradient(), StepLengthGradient(), Steps2PathinAtlasRRT(), Steps2PathinRRT(), Time2Go(), TransitionTestRRT(), TriangulateAtlas(), UpdateCuikSystem(), UpdateLQRPolicy(), WireAtlasRRTstar(), WireRRTstar(), WorldAtomJacobian(), WorldCoupleTensegrityVariable(), WorldDOF2Sol(), WorldForceField(), WorldForceVars(), WorldPotentialEnergy(), WorldSample2DOF(), and WorldSimpKinematicVars(). ◆ GetParameterID()
Determines the parameter identifier for a given parameter name.
Definition at line 118 of file parameters.c. References FALSE, NO_UINT, and NPARAMETERS. ◆ ParameterSet()
Determines is a parameter is already defined. The default initialization (the one from the default set of parameters) is not taken into account. Only explicit initializations from the users are considered. Only parameters alrady defined can be used in the definition of other parameters.
Definition at line 139 of file parameters.c. ◆ SetParameter()
Sets the name and value for a particular parameter.
Definition at line 147 of file parameters.c. References ChangeParameter(), Error(), NPARAMETERS, and Warning(). ◆ ChangeParameter()
Sets the value for a particular parameter.
Definition at line 172 of file parameters.c. References Error(), and NPARAMETERS. Referenced by AddVelocityEquations(), DealWithCP(), IncrementalSampleCuikSystemInBox(), InitAtlasFromPoint(), main(), MinimizeOnAtlas(), PathEffort(), SampleCuikSystemInBox(), SetParameter(), and SmoothSamples(). ◆ PrintParameters()
Writes a parameter set to a stream, that can be stdout. In principle the output of this function can be used as a parameter file (i.e., it can be parsed correctly with InitParametersFromFile).
Definition at line 189 of file parameters.c. References AMBIENT_SAMPLING, C_FCL, CT_CD_ENGINE, CT_CUT_X, CT_CUT_Y, CT_CUT_Z, CT_G_AXIS, CT_REPRESENTATION, CT_SAMPLING, CT_SPLIT_TYPE, Error(), FCL, INF, KDTREE_SAMPLING, NOCD, NPARAMETERS, PQP, REP_FLINKS, REP_JOINTS, REP_LINKS, REP_QLINKS, RIGIDCLL, SOLID, TANGENT_SAMPLING, and VCOLLIDE. Referenced by main(). ◆ DeleteParameters()
Deletes a set of parameters and frees the allocated memory.
Definition at line 361 of file parameters.c. Referenced by main(). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
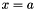 or
or 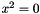 are considered (with x a variable and a a constant).
are considered (with x a variable and a a constant).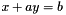 are also considered (with x and y variables and a and b constants).
are also considered (with x and y variables and a and b constants).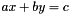 are also considered (with x and y variables and a and b constants).
are also considered (with x and y variables and a and b constants).
Follow us!